Arc Virtual Cell Challenge - part 4: Winners announced!
Did Bio-Foundation models outperform traditional statistical approaches?
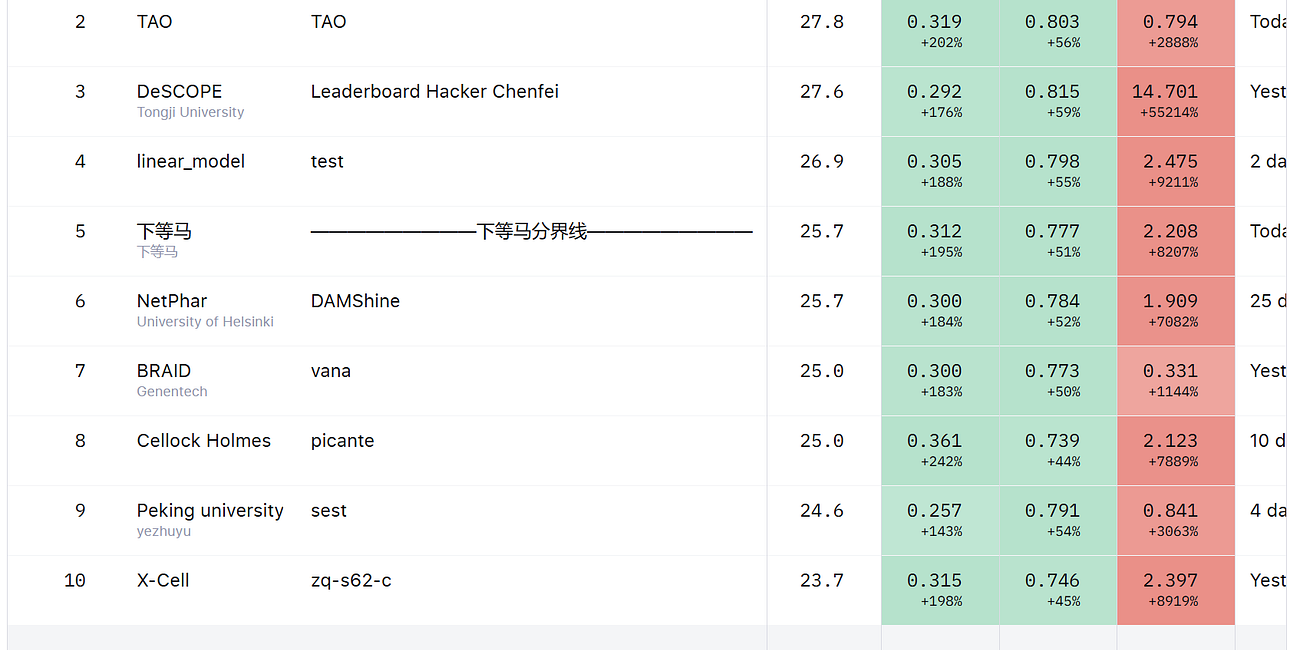
The Winners of the first Arc Virtual Cell Challenge have been announced! Congratulations to all the teams.
This wrap-up blog post on the Challenge’s website gives plenty of details about the winners and their strategies, along with some considerations on the challenge itself. Here are my thoughts.
The “leaderboard hacking” did not matter much in the end
It seems that the winning teams developed very robust and clever approaches. While the challenge highlighted the limitations of combining MAE, PBS and DE scores, this did not influence the final leaderboard. The winning teams did focus more on PBS and DES, and likely ignored MAE, but their models are still valid. It didn’t pay to overfit the predictions to the public validation set :-).
The organizers even awarded a new “Generalist Prize”, computed based on four additional metrics, mentioned in the STATE original paper. This award went to a team from Altos Labs.
In the Discord channel, one of the participants shared a nice article about leaderboard hacking. In their case, they focused on optimizing the DES scores, and understood that genes that are highly expressed in the controls are also likely to be Differentially expressed.
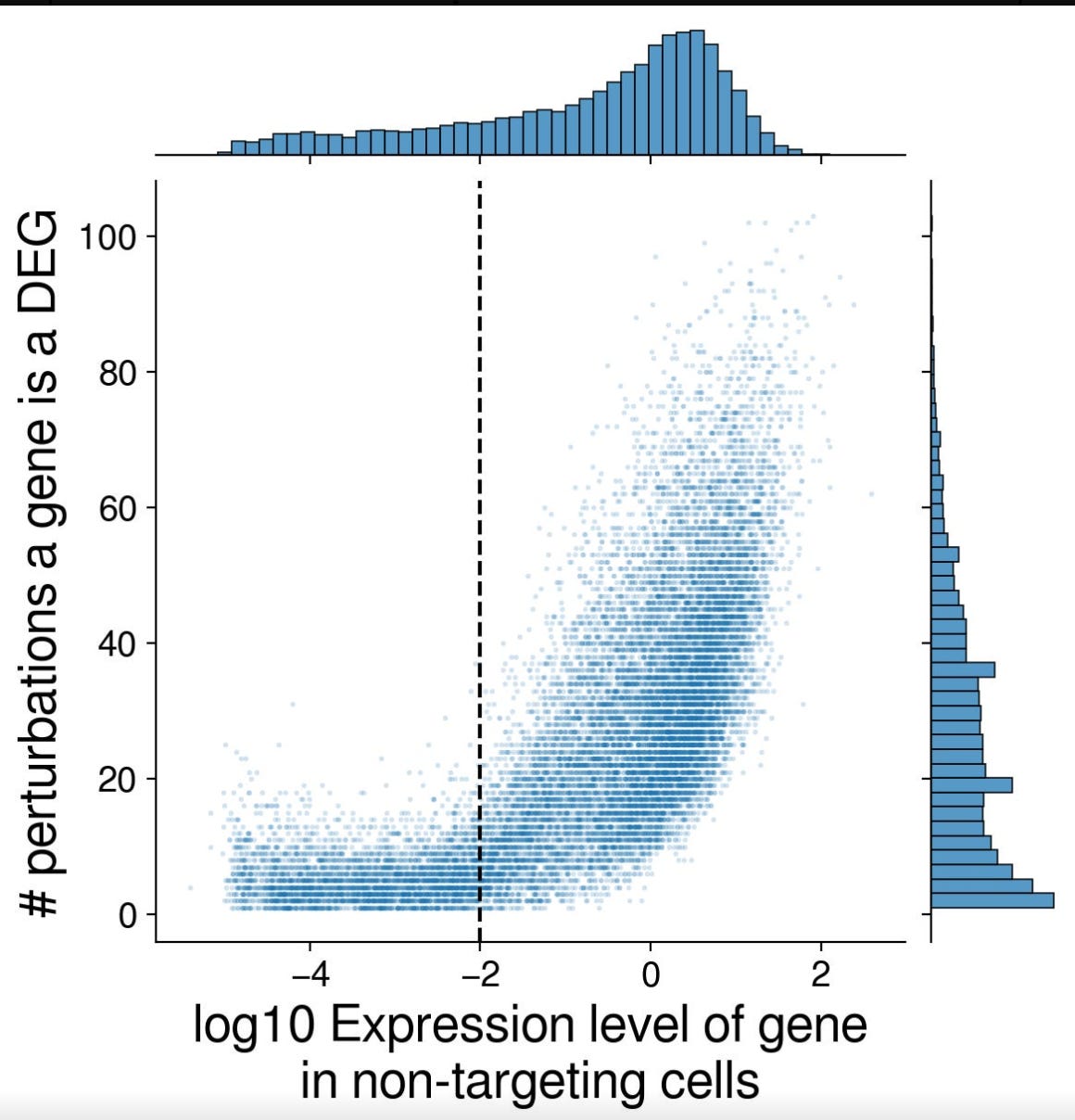
Personally, I think there is so much to learn from the efforts from hacking the challenge, than anything else. It highlights how difficult it is to evaluate these models in an objective way. As the authors of that article mention, it is difficult for a human to visualize a space with 18,000 dimensions, meaning that we need to develop biologically interpretable metrics to evaluate these models.
Did Bio-Foundation models out perform traditional methods? Or not?
In the run-up to the challenge, there was a debate in the scientific community about whether traditional methods, such as a simple regression, outperform these new big foundation models based on deep learning approaches.
Some of the main papers on this topic are "Zedzierska et al 2025: Zero-shot evaluation reveals limitations of single-cell foundation models” , and “Ahlmann-Eltze et al 2025: Deep-learning-based gene perturbation effect prediction does not yet outperform simple linear baselines”. Other papers, like “Miller et al 2025 Deep Learning-Based Genetic Perturbation Models Do Outperform Uninformative Baselines on Well-Calibrated Metrics”, responded by proposing better metrics to compare them.

Now that the challenge has finished and the winners announced, can we say whether this is true? Many teams competed in this challenge for over three months, some using traditional methods, and some using deep learning approaches. So, who got it right, in the end?
Looking at the winners of the challenge, I think that the answer is clear: hybrid approaches performed best. The winning team used a combination of deep learning, developing their own foundation model, and traditional statistics. They integrated previous knowledge on the effects of perturbations, inferring it from pseudo-bulk and literature, and used it to improve their predictions. Nobody won by using a regression alone, nor nobody won by using only STATE.
Pseudo-bulk is still important
Something that struck me from reading the article is that all the winning teams used pseudo-bulk in their strategies. If I read it right, the winners of the second and third place focused mostly on pseudo-bulk, while the top team integrated it into their model, as a way to correct for technical zero inflation.
This is not news to me. One of the participating teams shared a write-up, after the competition, about how they used a pseudo-bulk approach for this challenge. Not sure if the write-up can be shared publicly, but their approach is also described in this paper: Csendes et al, 2025 Benchmarking foundation cell models for post-perturbation RNA-seq prediction.
Is PBS a good metric for evaluating a Virtual Cell Model?
This challenge sparked a lot of discussion on the metrics to evaluate Virtual Cell perturbation models.
I just want to share this blog post, from my homonym Giovanni:
Another piece worth reading is this preprint from one of the participants: “Liu et al 2025, Effects of Distance Metrics and Scaling on the Perturbation Discrimination Score.”
What’s next?
There is much more to come.
First of all, I’ll be waiting to read the academic paper from the winning team, describing the details of the method.
This challenge was just the first, and the Arc institute promised one for the next year. I’ll be ready to participate!
Moreover, one of the participants is preparing a Kaggle challenge on this topic, possibly in January. This will be smaller, but focused on better evaluation metrics. I am looking forward to that.
My other posts on this challenge
Arc Virtual Cell Challenge part 1: my diary
The Arc Virtual Cell Challenge is a competition organized by the Arc Institute in the summer of 2025. The challenge is still ongoing, with the final deadline on November 17.
Arc Virtual Cell Challenge part 2: What the Leaderboard Collapse Reveals About Model Evaluation
The Arc Virtual Challenge is coming to an end, with the deadline in a mid November 2025.
Arc Virtual Cell Challenge part 3: how to train your own Virtual Cell Model
Here is my third article covering the Arc Virtual Cell Challenge (see first and second).