Arc Virtual Cell Challenge: my diary
A journey into bio-foundation models to predict perturbations in single-cell data
The Arc Virtual Cell Challenge is a competition organized by the Arc Institute in the summer of 2025. The challenge is still ongoing, with the final deadline on November 17.
I joined only recently - a bit late, due to work commitments - so I’m not expecting to win. My main goal is simply to learn. In this series of posts, I’ll share my experience and progress as I take part in the challenge.
What is the objective of the challenge?
The task is to predict the effect of gene perturbations in a human stem cell line known as H1.
In particular, the organizers only provide data from about 150 gene perturbations in H1 cell lines, plus some controls. The objective is to predict the effect of 50 gene perturbations in the same cell lines.
Any dataset or modelling approach can be used, but for most participants, the challenge serves as an opportunity to experiment with STATE, the large bio-foundation model released by the Arc Institute earlier this year.
A Possible Approach
This image, taken from a starter Google/Colab provided by the organizers, describes one possible approach to the challenge.
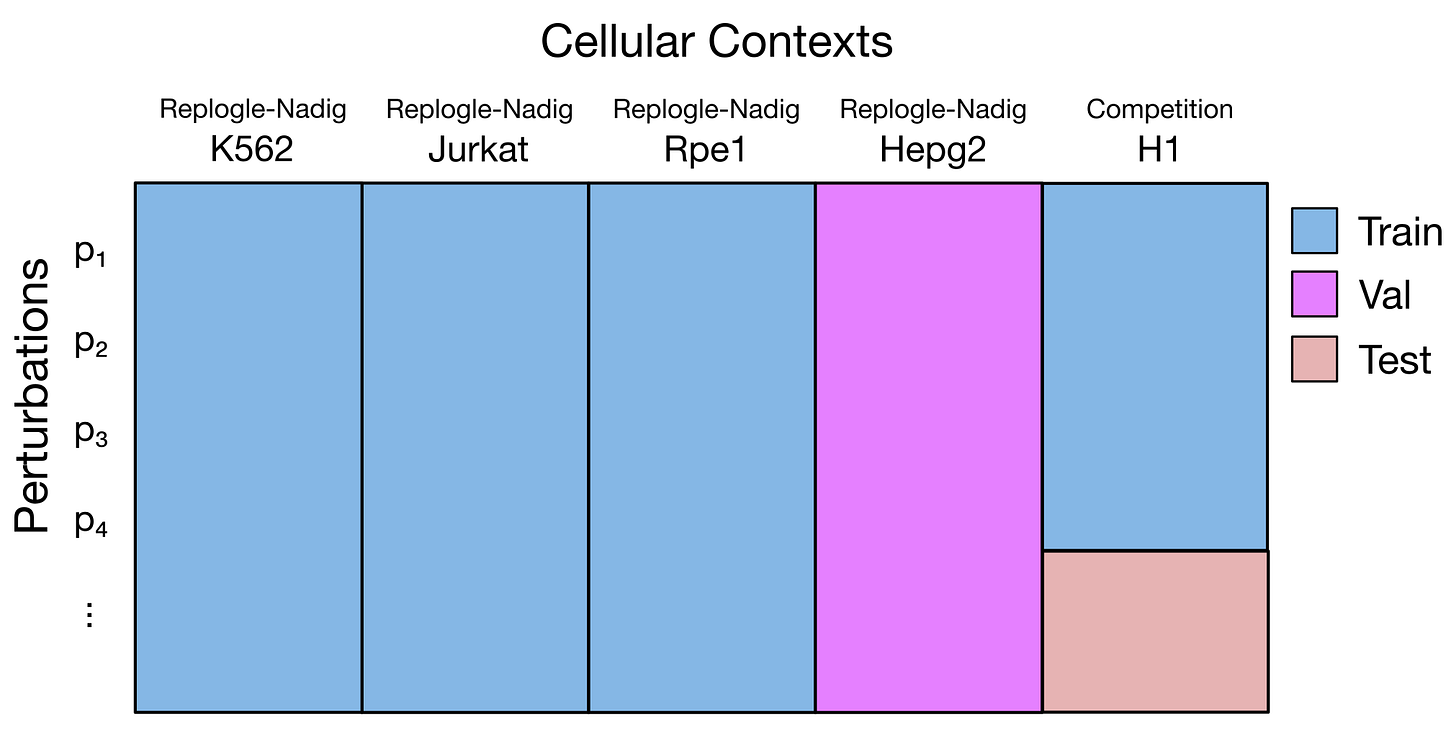
One possible approach is to augment the training data with other public datasets - such as the Replogle and Nadig datasets, which include perturbations of the same genes but in different cell lines.
We could train a machine learning model on these datasets to learn how each gene perturbation affects gene expression, and then use it to predict what would happen in H1. If perturbing gene X leads to lower expression of gene Y in most cell lines, we could assume that the same happens in H1 as well.
However, this comes with a major caveat: cell context matters. The H1 line is a stem cell line, while Replogle and Nadig involve cancer cell lines. The same perturbation can have drastically different effects depending on the cellular background.
To adjust for this, one idea would be to train a second model to learn the systematic differences between cancer lines and H1 - then apply those corrections to the predictions of the first model. This is an option, but somewhat complicated to put in practice.
Why Bio-Foundation Models Help
This is exactly where bio-foundation models like STATE shine.
Instead of training multiple models and manually engineering context corrections, we can train STATE on all available data - from Replogle and Nadig, plus the H1 training data.
The model then learns both the biological effects of gene perturbations and the contextual relationships between different cell types. Instead of implementing the two models described above, we create a single, big model that is able to generalize in both directions.
What’s Next
Over the next few weeks, I’ll document how I approach this challenge - from setting up the data to testing model variants and interpreting prediction.
I’ll go through the starter Colab Notebook, and document my progress in training STATE. It turns out to be easier than expected, but complex to tweak.
I’ll also go through the problem of validating the predictions, which for me, is one of the most interesting aspects of the challenge. Spoiler alert - there is a lot of discussion in the forums, because the metrics used for scoring are vulnerable to hacking.
If time allows, I’ll also cover other approaches alternative to STATE - from this Large Perturbation Model published recently, to simpler approaches.